Practice Phylogenetic Trees 1 Answer Key embarks on an enlightening journey into the realm of phylogenetic trees, unlocking the secrets of evolutionary relationships. Phylogenetic trees, intricate diagrams depicting the evolutionary history of species, serve as invaluable tools for understanding the diversity of life on Earth.
This comprehensive guide delves into the intricacies of phylogenetic tree construction, empowering readers with the knowledge and skills to navigate the complexities of evolutionary analysis. From grasping the basics to mastering advanced techniques, this answer key provides a roadmap for unraveling the mysteries of phylogenetic trees.
Phylogenetic Tree Basics: Practice Phylogenetic Trees 1 Answer Key
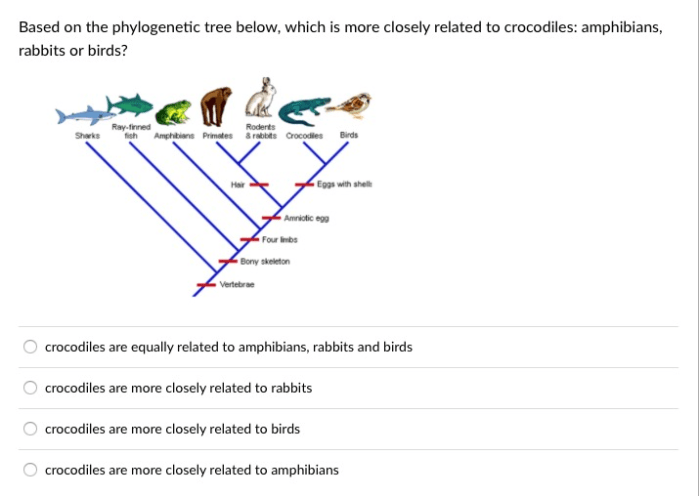
A phylogenetic tree, also known as an evolutionary tree or a branching diagram, is a diagrammatic representation that depicts the evolutionary relationships between different biological species or other taxa.
The purpose of a phylogenetic tree is to illustrate the evolutionary history and diversification of a group of organisms, showing their shared ancestry and the branching patterns that have led to their current diversity.
Different Types of Phylogenetic Trees, Practice phylogenetic trees 1 answer key
There are different types of phylogenetic trees, each based on specific criteria:
- Rooted treeshave a designated root node representing the common ancestor of all the taxa included in the tree.
- Unrooted treesdo not specify a root node, representing a hypothetical ancestor without assigning a specific direction to the branching.
- Cladistic treesrepresent evolutionary relationships based solely on shared derived characters (synapomorphies).
- Distance-based treesrepresent evolutionary relationships based on the amount of genetic or phenotypic divergence between taxa.
Practicing Phylogenetic Tree Construction
Practice phylogenetic trees are essential for developing an understanding of the principles and methods used in constructing and interpreting phylogenetic trees.
Here are some examples of practice phylogenetic trees:
- The Tree of Life, which depicts the evolutionary relationships of all living organisms.
- Phylogenies of specific taxonomic groups, such as the primate family tree or the evolutionary tree of dinosaurs.
- Trees that represent the relationships between different gene sequences or protein structures.
Methods for constructing phylogenetic trees include:
- Maximum parsimony: Assumes the simplest explanation for the observed data, minimizing the number of evolutionary changes required to explain the relationships.
- Maximum likelihood: Estimates the evolutionary tree that is most likely to have produced the observed data.
- Bayesian inference: Uses statistical methods to estimate the probability of different evolutionary trees given the observed data.
Tips for interpreting phylogenetic trees:
- Identify the root node, if present, which represents the common ancestor.
- Trace the branching patterns to understand the evolutionary relationships between different taxa.
- Pay attention to the branch lengths, which can indicate the amount of evolutionary change that has occurred along different lineages.
Key Considerations in Phylogenetic Tree Analysis
Data quality is crucial in phylogenetic tree analysis, as it directly affects the accuracy and reliability of the results.
Outgroups, which are taxa that are closely related to the ingroup but not included in the analysis, are used to root the tree and provide a reference point for determining the direction of evolutionary change.
Tree rooting establishes the ancestral state and allows for the identification of derived characters that support specific evolutionary relationships.
Advanced Techniques in Phylogenetic Tree Analysis
Estimating branch lengths provides insights into the rate of evolutionary change along different lineages.
Assessing tree reliability involves using statistical methods, such as bootstrapping and jackknifing, to evaluate the support for different branches in the tree.
| Method | Assumptions | Advantages | Disadvantages |
|---|---|---|---|
| Maximum Parsimony | Simplest explanation | Computationally efficient | May not always find the optimal tree |
| Maximum Likelihood | Most likely tree | Statistically rigorous | Computationally intensive |
| Bayesian Inference | Probabilistic approach | Considers uncertainty in the data | Computationally very intensive |
Applications of Phylogenetic Trees
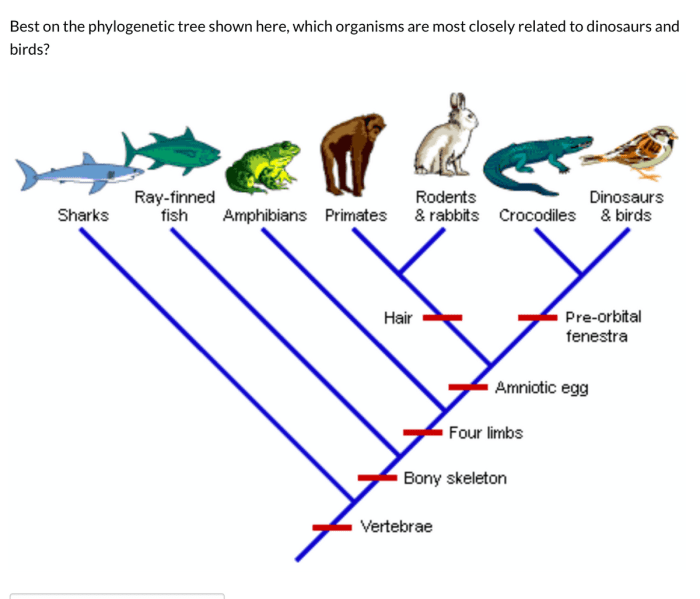
Phylogenetic trees are widely used in evolutionary biology:
- Understanding the evolutionary history of different species
- Inferring the timing of evolutionary events, such as speciation and extinction
- Identifying patterns of adaptation and diversification
In taxonomy, phylogenetic trees are used to:
- Classify organisms into different taxonomic groups
- Determine the evolutionary relationships between different species
- Identify new species and resolve taxonomic uncertainties
Phylogenetic trees also have applications in other fields, including:
- Biogeography: Studying the distribution of species and their evolutionary relationships
- Conservation biology: Identifying endangered species and prioritizing conservation efforts
- Medicine: Understanding the evolution of pathogens and developing new treatments
Frequently Asked Questions
What is the purpose of a phylogenetic tree?
Phylogenetic trees provide a visual representation of evolutionary relationships among different species, allowing scientists to infer their common ancestry and divergence over time.
How do I construct a phylogenetic tree?
Phylogenetic trees can be constructed using various methods, including maximum parsimony, neighbor-joining, and Bayesian inference. These methods analyze genetic or morphological data to infer the most likely evolutionary relationships.
What is the importance of data quality in phylogenetic analysis?
Data quality is crucial for accurate phylogenetic tree construction. Poor-quality data can lead to misleading or inaccurate trees, potentially compromising evolutionary interpretations.